SCIENTIFIC PUBLICATIONS
Here you can find all the scientific publications related to the Booster project
Authors: Lorena Ramirez-Gonzales, Gina Cannarozzi, Abiel Rindisbacher, Lea Jäggi, Regula Schneider, Annett Weichert, Sonia Plaza-Wüthrich, Solomon Chanyalew,Kebebew Assefa and Zerihun Tadele.
The threat to world food security posed by drought is ever increasing. Tef [Eragrostis tef (Zucc.) Trotter] is an allotetraploid cereal crop that is a staple food for a large population in the Horn of Africa. While the grain of tef provides quality food for humans, its straw is the most palatable and nutritious feed for livestock. In addition, the tef plant is resilient to several biotic and abiotic stresses, especially to drought, making it an ideal candidate to study the molecular mechanisms conferring these properties.
The transcriptome expression of tef leaf collected from plants grown under drought conditions was profiled using RNA-Seq and key genes were verified using RT-qPCR. This study revealed that tef exhibits a complex molecular network involving membrane receptors and transcription factors that regulate drought responses. We identified target genes related to hormones like ABA, auxin, and brassinosteroids and genes involved in antioxidant activity. The findings were compared to physiological measurements such as changes in stomatal conductance and contents of proline, chlorophyll and carotenoid. The insights gained from this work could play vital role in enhancing drought tolerance in other economically important cereals such as maize and rice.
Authors: Lorena Y. Ramírez Gonzales, Gina Cannarozzi, Lea Jäggi, Kebebew Assefa, Solomon Chanyalew, Matteo Dell’Acqua and Zerihun Tadele.
Tef research has little benefited from omics due to neglect of the crop by the global scientific community. However, the few utilized tools showed promising results. Chromosome-scale assembly of the genome reveals the evolution of tef. The whole-genome sequencing of drought-resurrecting Eragrostis nindensis and desiccation-sensitive Eragrostis curvula shows genes that play key roles in drought responses. A few studies using omics tools have revealed differentially expressed genes, proteins, and metabolites in tef plants exposed to drought. Molecular markers have deciphered diversity in tef germplasm collected from diverse agroecologies in Ethiopia. The application of targeting induced local lesions in genomes resulted in mutant tef lines with a semidwarf stature, which were later bred to locally adapted and high-yielding varieties. Genome editing using ‘Green Revolution’ genes resulted in a semidwarf and lodging-tolerant tef plant.
Tef or teff [Eragrostis tef (Zucc.) Trotter] is a cereal crop indigenous to the Horn of Africa, where it is a staple food for a large population. The popularity of tef arises from its resilience to environmental stresses and its nutritional value. For many years, tef has been considered an orphan crop, but recent research initiatives from across the globe are helping to unravel its undisclosed potential. Advanced omics tools and techniques have been directed toward the exploration of tef’s diversity with the aim of increasing its productivity. In this review, we report on the most recent advances in tef omics that brought the crop into the spotlight of international research.
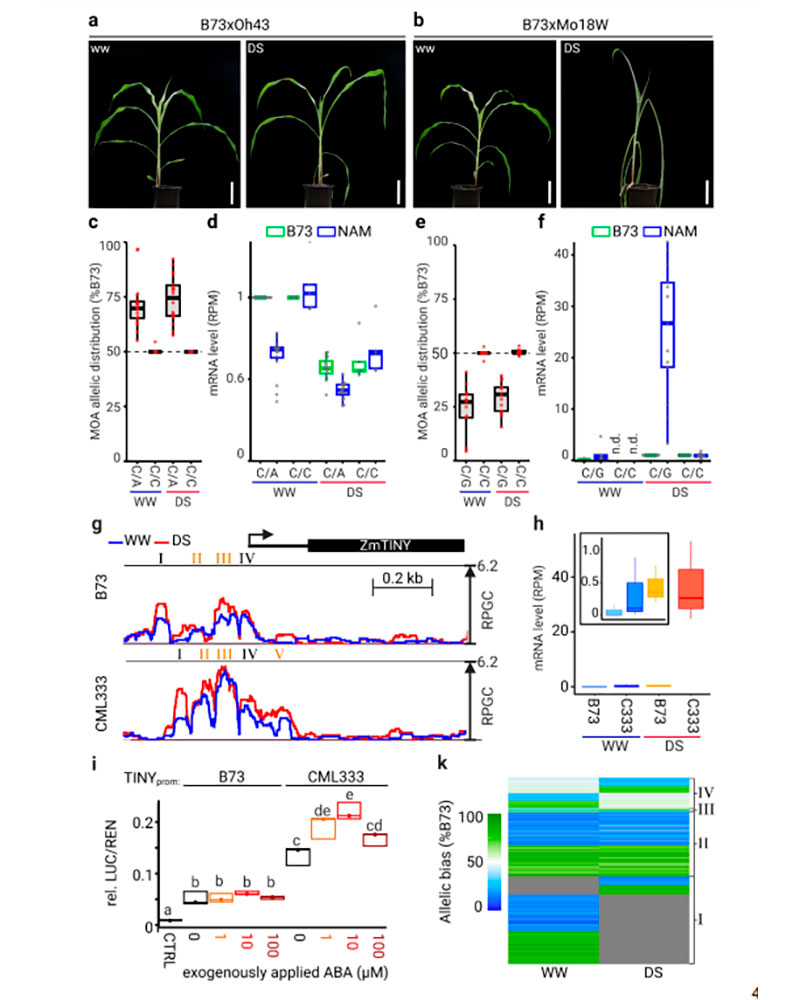
Authors: Julia Engelhorn, Samantha J. Snodgrass, Amelie Kok, Arun S. Seetharam, Michael Schneider, Tatjana Kiwit, Ayush Singh, Michael Banf, Merritt Khaipho-Burch, Daniel E. Runcie, Victor A. Sanchez-Camargo, J. Vladimir Torres-Rodriguez, Guangchao Sun, Maike Stam, Fabio Fiorani, Sebastian Beier, James C. Schnable, Hank W. Bass, Matthew B. Hufford, Benjamin Stich, Wolf B. Frommer, Jeffrey Ross-Ibarra and Thomas Hartwig.
Comprehensive maps of functional variation at transcription factor (TF) binding sites (cis-elements) are crucial for elucidating how genotype shapes phenotype. Here we report the construction of a pan-cistrome of the maize leaf under well-watered and drought conditions. We quantified haplotype-specific TF footprints across a pan-genome of 25 maize hybrids and mapped over two-hundred thousand genetic variants (termed binding-QTL) linked to cis-element occupancy. Three lines of evidence support the functional significance of binding-QTL: i) they coincide with numerous known causative loci that regulate traits, including VGT1, Trehalase1, and the MITE transposon near ZmNAC111 under drought; ii) their footprint bias is mirrored between inbred parents and by ChIP-seq; iii) partitioning genetic variation across genomic regions demonstrates that binding-QTL capture the majority of heritable trait variation across ∼70% of 143 phenotypes. Our study provides a promising approach to make previously hidden cis-variation more accessible for genetic studies and multi-target engineering of complex traits.
Authors: Nicolás Manosalva Pérez, Camilla Ferrari, Julia Engelhorn, Thomas Depuydt, Hilde Nelissen, Thomas Hartwig and Klaas Vandepoele.
Gene regulatory networks (GRNs) represent the interactions between transcription factors (TF) and their target genes. Plant GRNs control transcriptional programs involved in growth, development, and stress responses, ultimately affecting diverse agricultural traits. While recent developments in accessible chromatin (AC) profiling technologies make it possible to identify context-specific regulatory DNA, learning the underlying GRNs remains a major challenge. We developed MINI-AC (Motif-Informed Network Inference based on Accessible Chromatin), a method that combines AC data from bulk or single-cell experiments with TF binding site (TFBS) information to learn GRNs in plants. We benchmarked MINI-AC using bulk AC datasets from different Arabidopsis thaliana tissues and showed that it outperforms other methods to identify correct TFBS. In maize, a crop with a complex genome and abundant distal AC regions, MINI-AC successfully inferred leaf GRNs with experimentally confirmed, both proximal and distal, TF–target gene interactions. Furthermore, we showed that both AC regions and footprints are valid alternatives to infer AC-based GRNs with MINI-AC. Finally, we combined MINI-AC predictions from bulk and single-cell AC datasets to identify general and cell-type specific maize leaf regulators. Focusing on C4 metabolism, we identified diverse regulatory interactions in specialized cell types for this photosynthetic pathway. MINI-AC represents a powerful tool for inferring accurate AC-derived GRNs in plants and identifying known and novel candidate regulators, improving our understanding of gene regulation in plants.